Unregulated cell division is a hallmark of cancer, and one of the key proteins involved in controlling cell division is called FoxM1. Abnormal activation of FoxM1 is a common feature of cancer cells and is correlated with poor prognosis, metastasis, and resistance to chemotherapy.
Now researchers at UC Santa Cruz have determined the structure of this protein–a kind of “master switch” for cell division–in its inactive or “off” conformation. This new understanding of the structure of FoxM1 could ultimately be used to design new drugs that stabilize the protein in its inactive state and thereby stop the uncontrolled proliferation of cancer cells.
Seth Rubin, professor of chemistry and biochemistry at UC Santa Cruz, explained that FoxM1 is a “transcription factor,” a protein that controls the activity of specific genes.
“When a cell is going to divide, there are a bunch of proteins that need to be made, and FoxM1 controls all the genes for those proteins,” Rubin said. “Because cancer cells are proliferating and dividing all the time, they need to activate FoxM1, so it has long been a target for drug development.”
The new study involved a close collaboration between Rubin’s lab and that of Nikolaos Sgourakis, assistant professor of chemistry and biochemistry. After determining the structure of the protein in the “off” state, the team then figured out how it switches from the inhibited conformation to the activated or “on” state. The researchers published their findings May 28 in the journal eLife.
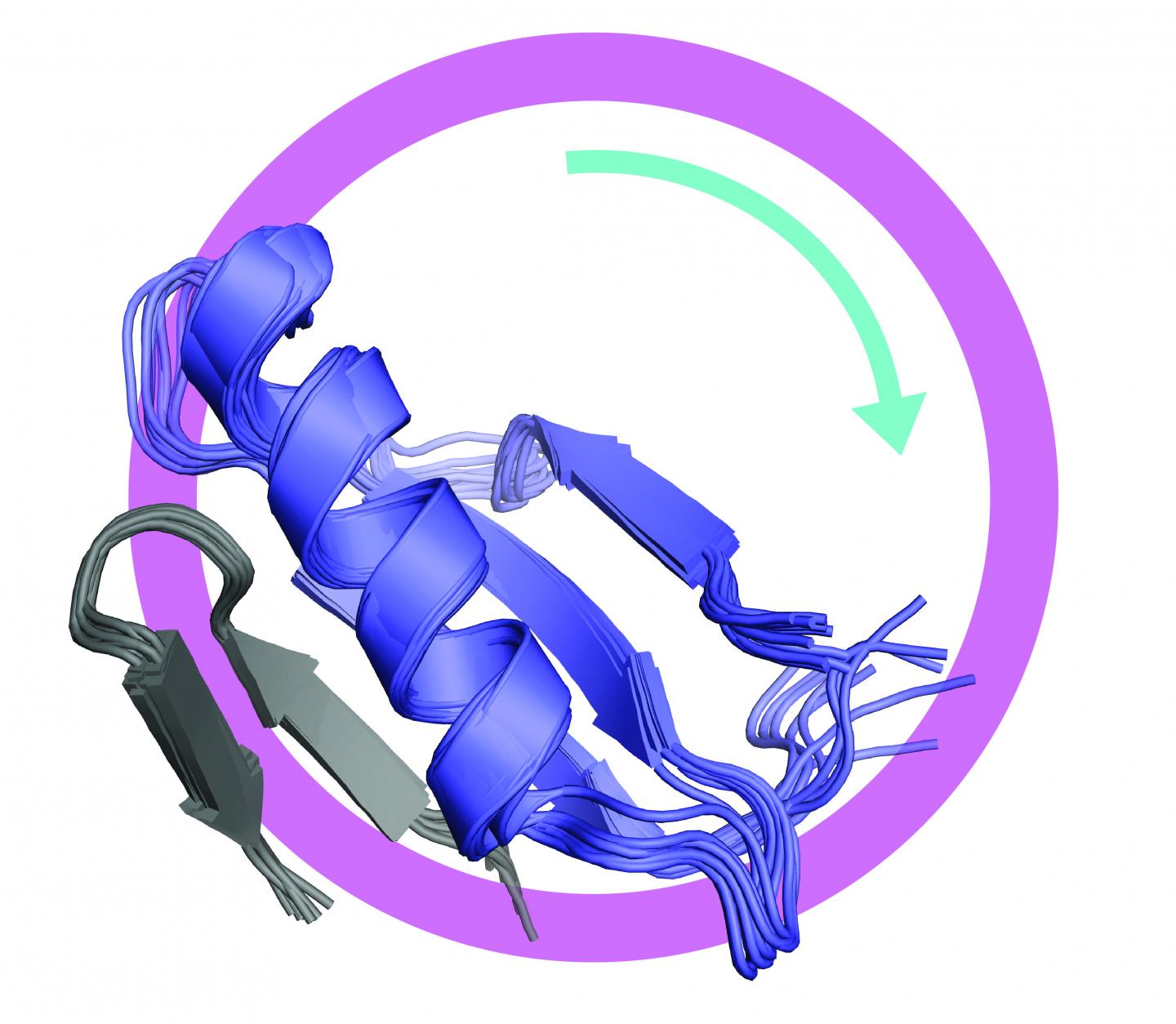
The study revealed that two separate domains of the FoxM1 protein interact and bind together in the inhibited conformation. The study also showed that the two domains separate and lose their structure when the protein is activated. Most proteins fold into an orderly three-dimensional structure that is key to their function, but some proteins function as disordered linear molecules with no particular 3D structure.
“One thing a disordered state is good at is interacting with other proteins,” Rubin said. “With FoxM1, the inactive state is all folded up on itself. When it gets activated it becomes disordered, and then it can recruit the other proteins needed to turn on gene expression. That’s something that hasn’t been seen before, and it may be a general mechanism for how transcription factors switch from the off state to the active state.”
It was known from previous studies that FoxM1 is activated by kinase enzymes, which add phosphoryl groups to specific sites on the protein. Rubin’s team found that phosphorylation of FoxM1 at one particular site causes the dissociation of the two domains and that both domains then become structurally disordered.
“Knowing the structure of the inhibited state of the protein really opens up a pathway to search for compounds that can stabilize it,” Rubin said. “And beyond drug development, in terms of the understanding of how transcription factors work, the discovery of this transition from an ordered to a disordered state is an important advance.”
The determination of the structure was not easy, he said. The researchers used a technique called NMR spectroscopy, taking advantage of a powerful new instrument at the UC Santa Cruz NMR Facility. Rubin’s initial work on FoxM1 was funded by a grant from the Santa Cruz Cancer Benefit Group. The new study was supported by grants from the National Institutes of Health, the American Cancer Society, and Alex’s Lemonade Stand Foundation.
###
In addition to Rubin and Sgourakis, the authors of the paper include co-first authors Aimee Marceau, Caileen Brison, and Santrupti Nerli at UC Santa Cruz, Heather Arsenault and Jennifer Benanti at University of Massachusetts Medical School, and Andrew McShan, Eefei Chen, and Hsiau-Wei Lee at UC Santa Cruz.
Disclaimer: AAAS and EurekAlert! are not responsible for the accuracy of news releases posted to EurekAlert! by contributing institutions or for the use of any information through the EurekAlert system.

