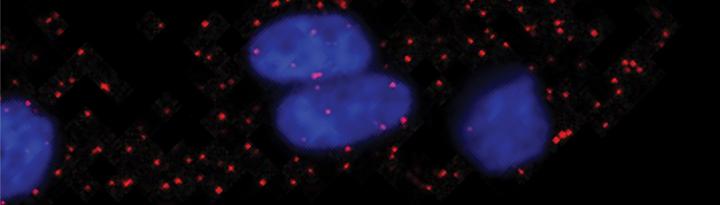
IMAGE: The breast cancer cells’ nuclei are illuminated (blue) by quantum dots and individual EGF growth factors appear as red spots. view more
Credit: University of Illinois Department of Bioengineering
Whether healthy or diseased, human cells exhibit behaviors and processes that are largely dictated by growth factor molecules, which bind to receptors on the cells. For example, growth factors tell the cells to divide, move, and when to die–a process known as apoptosis.
When growth factor levels are too high or too low, or when cells respond irregularly to their directions, many diseases can result, including cancer. “It is believed that cells respond to growth factors at extreme levels of sensitivity,” said University of Illinois at Urbana-Champaign Bioengineering Associate Professor Andrew Smith. “For example, a single molecule will result in a major change in cell behavior.”
In a recent paper published in Nature Communications, Smith reported the invention of a new technology platform that digitally counts, for the first time ever, the amount of growth factor entering an individual cell. Prior to this, researchers inferred growth factor binding based on how the receiving cells responded when the growth factor molecules were introduced.
“We showed the first direct cause-and-effect relationships of growth factors in single cells,” he said. “We expect the outcomes to lead to a new understanding of cell signaling, how cells respond to drugs, and why cell populations become resistant to drugs, particularly toward improved treatments for cancer.”
Smith’s technology platform tags each growth factor with a single engineered (10 nanometer) infrared fluorescent quantum dot, which can then be viewed using a three-dimensional microscope. In their study, they counted how many epidermal growth factor (EGF) molecules bound to human triple-negative breast cancer cells that were pre-patterned on island-like surfaces.
EGF molecules typically signal cell division and lead to tissue growth. Numerous cancers have mutations in their EGF receptors.
“We used quantum dots as the fluorescent probe because they emit a lot more light compared to other conventional fluorescent probes such as organic dyes, and we can tune their wavelengths by changing their chemical composition,” said Bioengineering doctoral student Phuong Le, the lead author of the paper. “In our study, we demonstrated that quantum dots emitting light in the near-infrared wavelength allowed the most accurate counting of growth factors binding to cells.”
According to Le, the team also treated the breast cancer cells with quantum dot-tagged EGF in the absence and presence of pharmaceutical drugs that inhibit EGF signaling in cells. “We found that the amount of EGF binding is inversely proportional to drug efficacy,” Le said. “This finding is significant as it means that signaling molecules present in the cancer cells’ tumor–a place where signaling molecules are often misregulated–can enhance the cancer cells’ resistance to pharmaceutical agents.”
###
In addition to Smith and Le, other research contributors to this study include Illinois Molecular and Integrative Physiology Professor Hee Jung Chung and graduate student Brian Baculis, who conducted molecular biology confirmation studies; former Illinois Postdoctoral Researcher Sung Jun Lim, who performed quantum dot synthesis; and University of New South Wales Professor Kristopher Kilian, who designed the micro contact printing process for the cell islands.
This work was funded by the National Institutes of Health and the University of Illinois at Urbana-Champaign.
Recently, Smith and Bioengineering Assistant Professor Pablo Perez-Pinera received more than $1 million in funding from the National Institutes of Health to further expand Smith’s novel technology with new cell engineering tools and image analysis software. The goal of their R01 grant will be to develop a quantitative analysis platform for single-cell signaling through growth factors and cytokines.
Disclaimer: AAAS and EurekAlert! are not responsible for the accuracy of news releases posted to EurekAlert! by contributing institutions or for the use of any information through the EurekAlert system.

