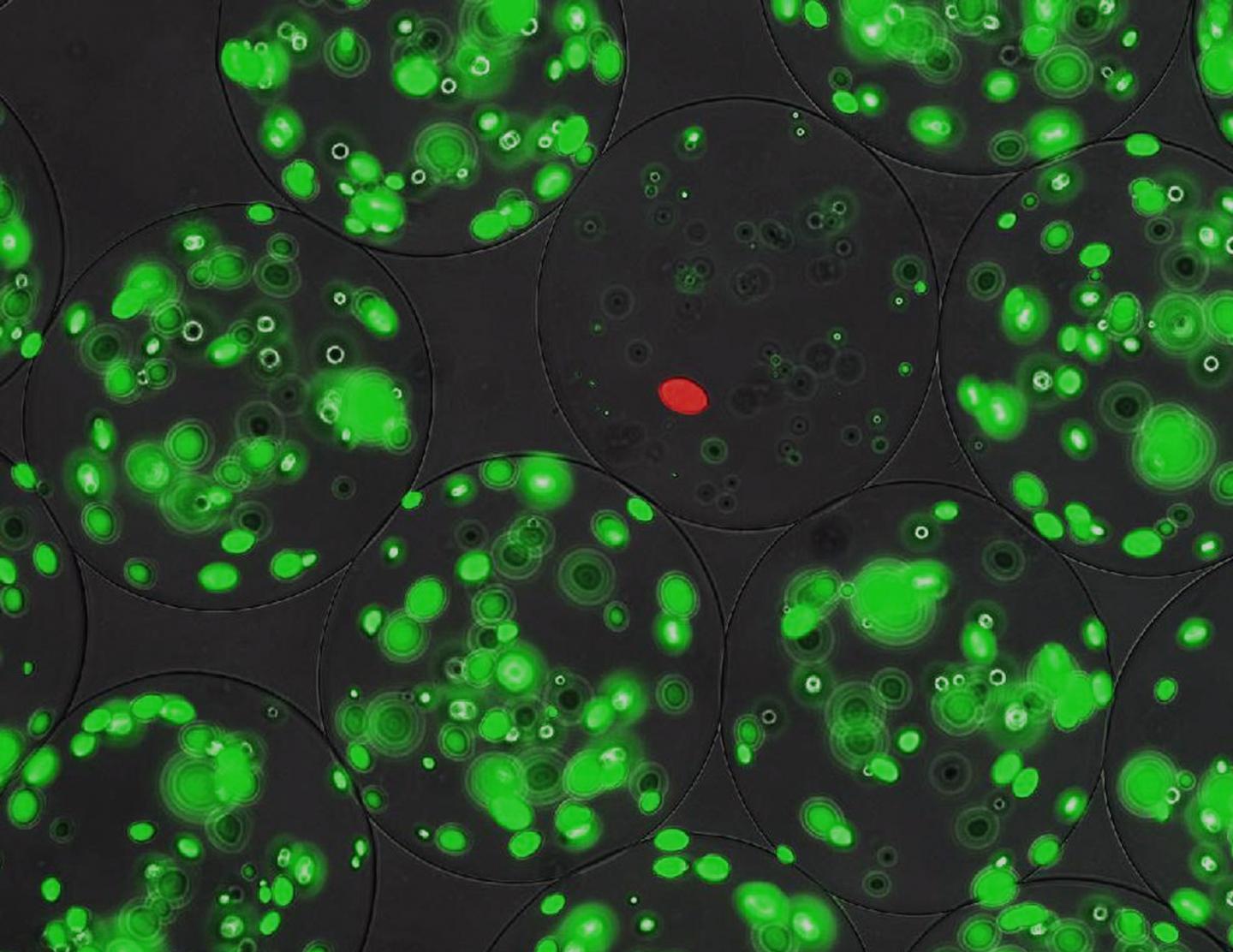
IMAGE: Alginate microbeads containing colonies of target cells (green) and producer cells (red). Where the producer cells indeed secrete active lantibiotics, the target cells have disappeared. view more
Credit: Steven Schmitt ETH Zürich
Nature has provided us with lots of antimicrobials. However, given the rapid increase of antimicrobial resistance, there is a need for the development of new-to-nature antibiotics. Lantibiotics are an interesting option. Molecular biologists from the University of Groningen and their colleagues in Switzerland and Germany have now developed a pipeline to create and screen large numbers of new lantibiotic peptides. A description of the method and the first results were published on 1 April in the journal Nature Chemical Biology.
Lantibiotic peptides are modified after they are produced by ribosomes. Enzymes can link different amino acids within the peptide chain to form rings. A well-known lantibiotic is nisin, a peptide with five rings that is used as a preservative in the food industry. ‘There are different amino acid sequences that make up the rings,’ explains University of Groningen Professor of Molecular Biology Oscar Kuipers. ‘We know that a selection of 12 natural lantibiotics all have different combinations.’
Synthetic genes
Kuipers and his colleagues from the ETH Zürich (Switzerland) and University of Regensburg (Germany) devised a system to create large numbers of new lantibiotics: ‘We synthesized DNA strands coding for the different rings and combined these genetic modules to form lantibiotic genes coding for random combinations of five rings.’ The gene library that they created contained around 14,000 synthetic genes.
The next step was to screen the gene products for antimicrobial potential. To do this, they modified a technique developed for enzyme screening, based on micro-alginate beads. Inside these beads – each around 70 micrometers in diameter – bacteria can grow. The genes from the library were put into a producer strain, containing a red fluorescent marker. ‘We diluted these bacteria so that we would end up with one or zero cells per bead. We also added a target strain that could be killed by the lantibiotics; those cells were used at a higher concentration of around 50 per bead.’ The target cells carried a green fluorescent marker and produced a peptidase that activated the precursor lantibiotics secreted by the producer cells.
Screening
When the beads were incubated, the producer strain formed a single colony inside, secreting lantibiotics. These peptides could not leave the beads. The number of green colonies growing inside each bead was negatively correlated with the antimicrobial effect of the activated lanthipeptides. Using a fluorescence-assisted cell sorter designed for particles, it was possible to screen 50,000 beads per hour and to select beads with low levels of green fluorescence.
But this screening was by no means the end of the work that needed to be done. The next step was to isolate producer colonies from the selected beads that showed a strong antimicrobial effect. This was straightforward; however, the exact characterization of the product formed turned out to be difficult because of the low production levels inside the bead. ‘We first had to grow them in larger quantities before we could analyze the lantibiotic produced.’ Another complication was that the enzymes producing the rings didn’t work with perfect accuracy. ‘Even in one colony, where all cells have the same DNA, up to five or six different lanthipeptides could be produced.’
Rational design
In the end, a series of lanthipeptides with a strong antimicrobial effect was isolated and characterized. ‘We found peptides with different antibiotic activities, although several peptides looked a bit like nisin or epidermin. It turned out that we had even recreated nisin in our gene library.’ The experiment provided a lot of information about the structure-function relationship of the different rings in the lantibiotic peptides. This can now be used for a rational design approach to create new antibiotics.
A second conclusion from the study is that the pipeline – from producing a gene library to screening and characterization – works very efficiently. The next step is to use a real pathogen as a reporter strain, instead of the Lactococcus cells used in the first round. ‘And we are now working to create peptides with just one ring. The five-ring peptides that we produced in this study are too large for pharmaceutical use. Many antibiotics resemble a one-ring structure, similar to that of vancomycin.’ These antibiotics are made by specific enzymatic linkage of amino acids, whereas the lantibiotics are produced from genes by ribosomes. ‘This means that we can use rapid molecular biology techniques to create and screen large libraries.’
###
Reference: Steven Schmitt, Manuel Montalbán-López, David Peterhoff, Jingjing Deng, Ralf Wagner, Martin Held, Oscar P. Kuipers & Sven Panke: Analysis of modular bioengineered antimicrobial lanthipeptides at nanoliter scale. Nature Chemical Biology 1 April 2019
Disclaimer: AAAS and EurekAlert! are not responsible for the accuracy of news releases posted to EurekAlert! by contributing institutions or for the use of any information through the EurekAlert system.

