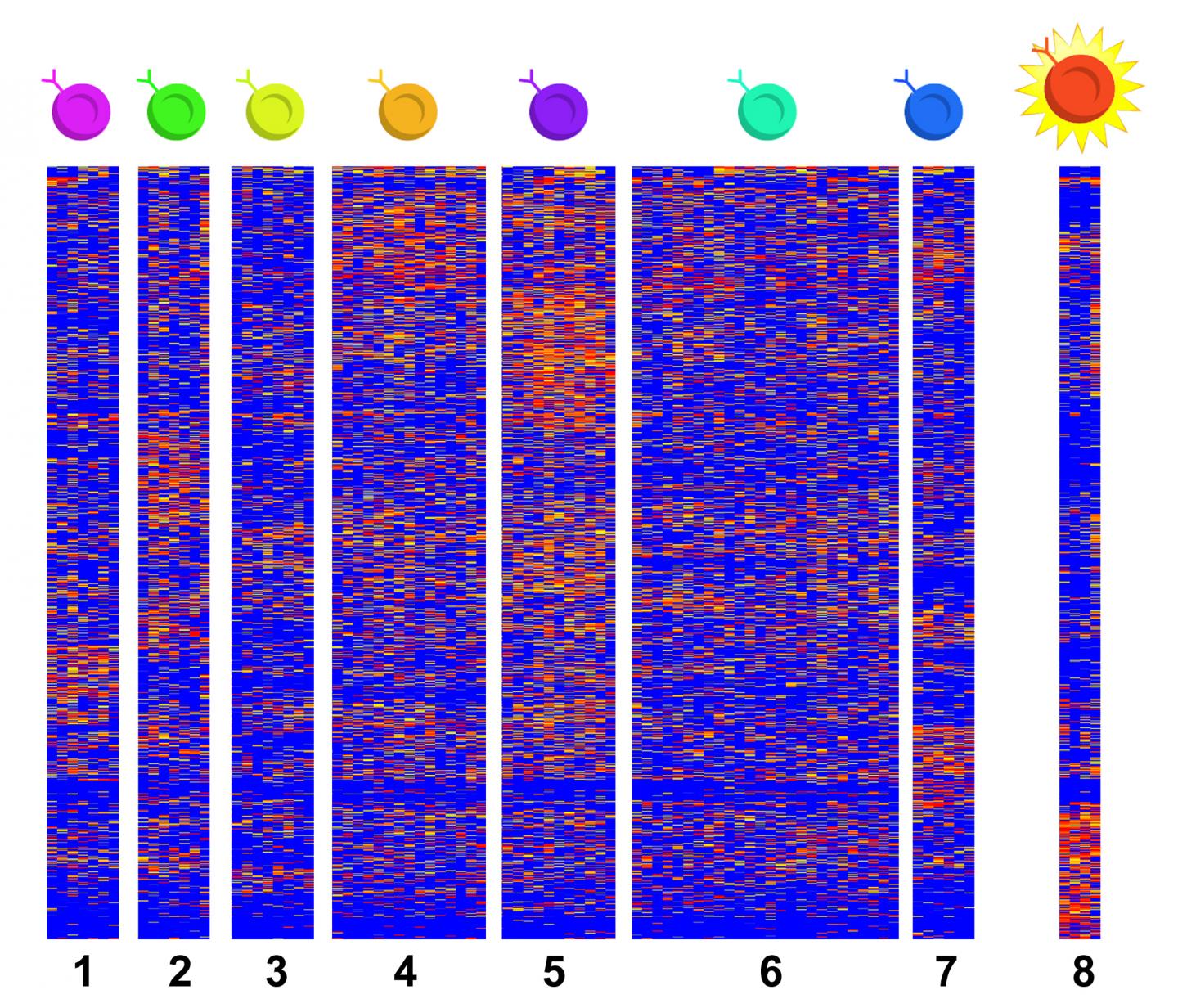
IMAGE: This heat map shows single-cell RNA sequencing data from eight types of tissue-resident T cells found in tissues collected from people with eosinophilic esophagitis. T8 cells are not found in… view more
Credit: Cincinnati Children’s
CINCINNATI–An eight-year hunt for the cells that drive the extreme childhood food allergy eosinophilic esophagitis (EoE) has identified a potential new way to treat the disease while also raising questions about a dietary supplement often taken to reduce bowel inflammation.
The study, led by experts at Cincinnati Children’s Hospital Medical Center, is posted online now in the Journal of Clinical Investigation and will be published in print in May. The work was led by first author Ting Wen, MD, PhD, and senior author Marc Rothenberg, MD, PhD, Director of the Division of Allergy and Immunology.
Reported cases of EoE have risen sharply in the past two decades, now affecting as many as one of every 2,000 people. This lifelong allergic disease occurs when too many eosinophils (a type of white blood cell) cause inflammation that makes swallowing and eating painful. EoE can be so serious that some people depend upon special liquid formulas to eat.
Rothenberg’s work over the years has helped establish EoE as its own disorder while Cincinnati Children’s has grown to become the nation’s leading center for treating and studying the disease. Clinical teams here help patients achieve better levels of remission while lab teams chase down the genetic and molecular pathways that eventually may lead to even better medications.
In this study, Wen and Rothenberg developed and perfected a 10- to 12-hour process that isolates and analyzes individual living cells from human tissue samples about the size of three sesame seeds.
“This is actually a substantial advance for the field,” Rothenberg says. “Prior work has focused on blood cells, but not the specific memory immune cells in tissue that directly respond to food allergens. We uncovered the unknown properties of these cells.”
Their key findings:
- Eight types of immune system T cells were found in the esophageal tissue. That’s more than had been previously expected.
- Two cell types (T7 and T8) show up in higher numbers in the diseased tissue. The study finds that one of these two cell types responds to allergens by cranking out massive amounts of inflammation-causing type 2 cytokines.
- One of these cell types (T8) is nearly non-existent in healthy tissue, making it especially worth targeting for treatment. “These cells are bad actors that should not be in this type of tissue. Inhibiting these cells is now a central goal of our research,” Wen says.
- So how did these bad players get there? Studying this question led the co-authors to a surprising finding. One of the overactive genes that helps trigger the inflammation response–a gene called FFAR3–also encodes a receptor for a dietary supplement called butyrate.
Good for the Lower Digestive Tract, Not so Much for the Upper
Butyrate is a short-chain fatty acid sold online and in many health food stores, without need for prescription. In the colon, this fatty acid helps the body produce energy from dietary fiber. It also appears to prevent or reduce inflammation in the lower digestive system, which has made the supplement popular among people battling inflammatory bowel diseases.
But in the esophagus, at the upper end of the digestive system, this otherwise beneficial fatty acid may drive or aggravate EoE by activating the newly identified population of cytokine-producing T cells, the co-authors say.
More studies are needed to answer questions such as: Are EoE patients aggravating their own conditions by taking butyrate? Does taking butyrate for a lower digestive tract problem create a higher risk of developing EoE? Do butyrate supplements need warning labels?
“It is far too early to say under which or any conditions butyrate could be harmful,” Rothenberg says. “But this initial discovery deserves further exploration.”
A Race for Results
A growing number of scientists are using single-cell RNA technology to study various diseases. This is the first study to use single-cell RNA sequencing to glean data from cells hiding within EoE-affected tissue.
This project required arranging for study participants to arrive early in the morning for doctors to remove a flake of tissue about the size of three sesame seeds. These samples were rushed to the lab where Wen painstakingly isolated more than 1,000 targeted cells, each suspended in its own microchamber on a special plastic chip. Then the team used RNA sequencing to profile their entire genomes, cell by cell by cell.
Each patient donor launched a day-long cell processing marathon that involved as many as 19 professionals. “There were days when people literally ran with the samples from the clinic to our laboratory. Many times, people agreed to work late into the evening to complete the process,” Wen says. “This truly was team science in action.”
Method May Also Help Study Asthma and Other Conditions
Beyond EoE, the process used in this study could be applied in other, more-common conditions to find small populations of cellular culprits hiding within tissues. The study co-authors say the process could be useful in studying asthma, atopic dermatitis, allergic rhinitis, and other diseases associated with type 2 cytokines.
“I have already been contacted by other labs interested in using this platform to study tissue-resident cells,” Wen says.
###
This study was supported by the National Institutes of Health (P30 DK078392, U19 AI070235, R01 AI124355, R37 A1045898); the HOPE Research Award (APFED); the Sunshine Charitable Foundation, and its supporters Denise and David Bunning; and by long-term support of the CURED Foundation.
A patent application related to the FFAR3 and T8 cell discoveries has been filed. Rothenberg and Wen also are listed as inventors on other patents held by Cincinnati Children’s. Rothenberg’s consulting and equity roles also are disclosed in the JCI article.
Disclaimer: AAAS and EurekAlert! are not responsible for the accuracy of news releases posted to EurekAlert! by contributing institutions or for the use of any information through the EurekAlert system.

