IMAGE: Aging doubles your risks of chronic diseases every eight years and there are genes responsible for that. view more
Credit: Gero LLC
The global population aged 60 or over is growing faster than all younger age groups and faces the tide of chronic diseases threatening their quality of life and posing challenges to healthcare and economy systems. To better understand the underlying biology behind healthspan — the healthy period of life before the first chronic disease manifestation — the scientists from Gero and MIPT collaborated with the researchers from PolyOmica, the University of Edinburgh and other institutes to analyze genetic data and medical histories of over 300,000 people aged 37 to 73 made available by UK Biobank.
The study published today in Communications Biology was lead by Dr. Peter Fedichev and Prof. Yurii Aulchenko. It shows that the most prevalent chronic conditions such as cancer, diabetes, chronic obstructive pulmonary disease, stroke, dementia, and some others apparently share the common underlying mechanism that is aging itself.
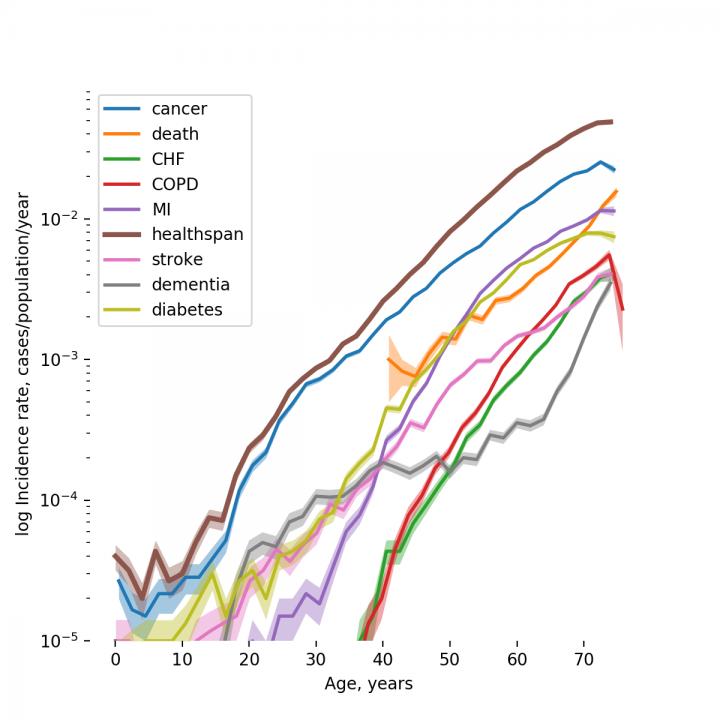
«According to Gompertz mortality law, the risk of death from all causes increases exponentially after the age of 40 and doubles approximately every 8 years», explains Peter Fedichev, founder and CSO of Gero. «By analyzing the dynamics of disease incidence in the clinical data available from UKB, we observed that the risks of age-related diseases grow exponentially with age and double at a rate compatible with the Gompertz mortality law. This close relation between the most prevalent chronic diseases and mortality suggests that their risks could be driven by the same process, that is aging. This is why healthspan can be used as a natural proxy for investigation of the genetic factors controlling the rate of aging, the “holy grail” target for anti-aging interventions».
To find out genetic factors associated with human healthspan, the researchers studied the genomes of 300,477 British individuals. Overall, 12 genetic loci affecting healthy life expectancy were discovered. To confirm that these results hold true for other ethnicities, they used genetic data of UK Biobank participants with self-reported European, African, South Asian, Chinese and Caribbean ancestry. Of the 12 SNPs, 11 increased risk both in discovery and in replication groups. Three of the genes affecting healthspan, HLA-DBQ, LPA, and CDKN2B, were previously associated with parental longevity, a proxy for overall life expectancy.
At least three genetic loci were associated with risk of multiple diseases and healthspan at the same time and therefore could form the genetic signature of aging. HLA-DQB1 was significantly associated with COPD, diabetes, cancer, and dementia in this study and was demonstrated to be associated with parental survival earlier. The genetic variants near TYR predict death in the prospective UKB cohort and are involved in the earlier onset of macular degeneration. The chromosome 20 locus containing C20orf112 was not associated with the incidence of any of the diseases at the full-genome level, and yet was affecting the healthspan of studied individuals.
Five healthspan-associated SNPs were also associated with a number of complex traits such as skin cancer, color of skin, eye and hair, tanning and freckles, coronary artery disease, myocardial infarction, cholesterol and glucose levels, BMI and type 2 diabetes. The researchers also found strong genetic correlations between healthspan and conditions such as obesity, type 2 diabetes, coronary artery disease, and sociodemographic factors including parental age at death, smoking and education level.
The studies of longevity genetics are complicated due to the limited availability of datasets containing clinical data of genotyped individuals who already reached the end of lifespan. Healthspan as a target phenotype offers a promising new way to interrogate the genetics of human longevity by untapping the research potential of large cohorts of living individuals with rich clinical information such as UK Biobank.
«The inevitable aging of global population calls for longevity research to focus on understanding the pathways controlling healthspan. Our study highlights the potential of GWAS methodology to delineate genetic architectures of healthspan and lifespan. We hope that our work will eventually help to produce novel diagnostic tools in the field of genetics of aging and to shape the target space for future therapeutics against aging. At the end of the day, healthspan, not lifespan, is the ultimate goal of future life-extending interventions!», concludes Peter Fedichev, the founder and Chief Scientific Officer of Gero.
###
About Gero
Gero is a data-driven longevity company developing innovative therapies that will strongly extend the healthy period of life also known as healthspan. Having a pipeline of longevity therapeutics in development, Gero is also providing health and mortality risk assessment models for life & health insurance, healthcare, and wellness industries.
Gero R&D team led by Dr. Peter Fedichev applies methods from dynamics systems and physical kinetics to produce predictive models of aging process for anti-aging targets and aging biomarkers identification. Recent results include experimental therapies reducing biological age in mice, newly identified compounds extending life in other model animals, and application of deep convolutional neural networks for identification of biomarkers of aging and frailty from wearable devices. The team has also introduced free iOS application for healthspan prediction from intraday physical activity of smartphone users.
Gero is a partner of Longevity Therapeutics 2019 (January 29-31, San Francisco) that will bring together the leading biotech drug developers, academics, investors and pharma companies endeavoring to develop innovative therapies targeting age-related conditions. Peter Fedichev’s talk «Hacking Aging: A Data-Driven Approach to Healthy Life Extension» will open the session «Supporting Discovery in Aging Research With Big Data» on January 30, 2019. Meet us there.
Disclaimer: AAAS and EurekAlert! are not responsible for the accuracy of news releases posted to EurekAlert! by contributing institutions or for the use of any information through the EurekAlert system.

